Running the ab initio reconstruction¶
Select the ab initio reconstruction. This protocol is easy to use. Simply select the extracted particles and the PSF you just imported.
In this protocol you can change the Expert Level to Advanced. It allows you to modify more parameters, especially in the Reconstruction params tab.
Click on  to get explanations.
to get explanations.
This protocol will take some time to finish. When executed, a tab in your browser will be opened. It plots the energy after each epochs.
To see if the run is doing well, go to the Output Log tab on the bottom panel.
Here you have run.stdout which prints all the commands that were called during the run;
and, more importantly, run.stderr which gives you the progression of the algorithm.
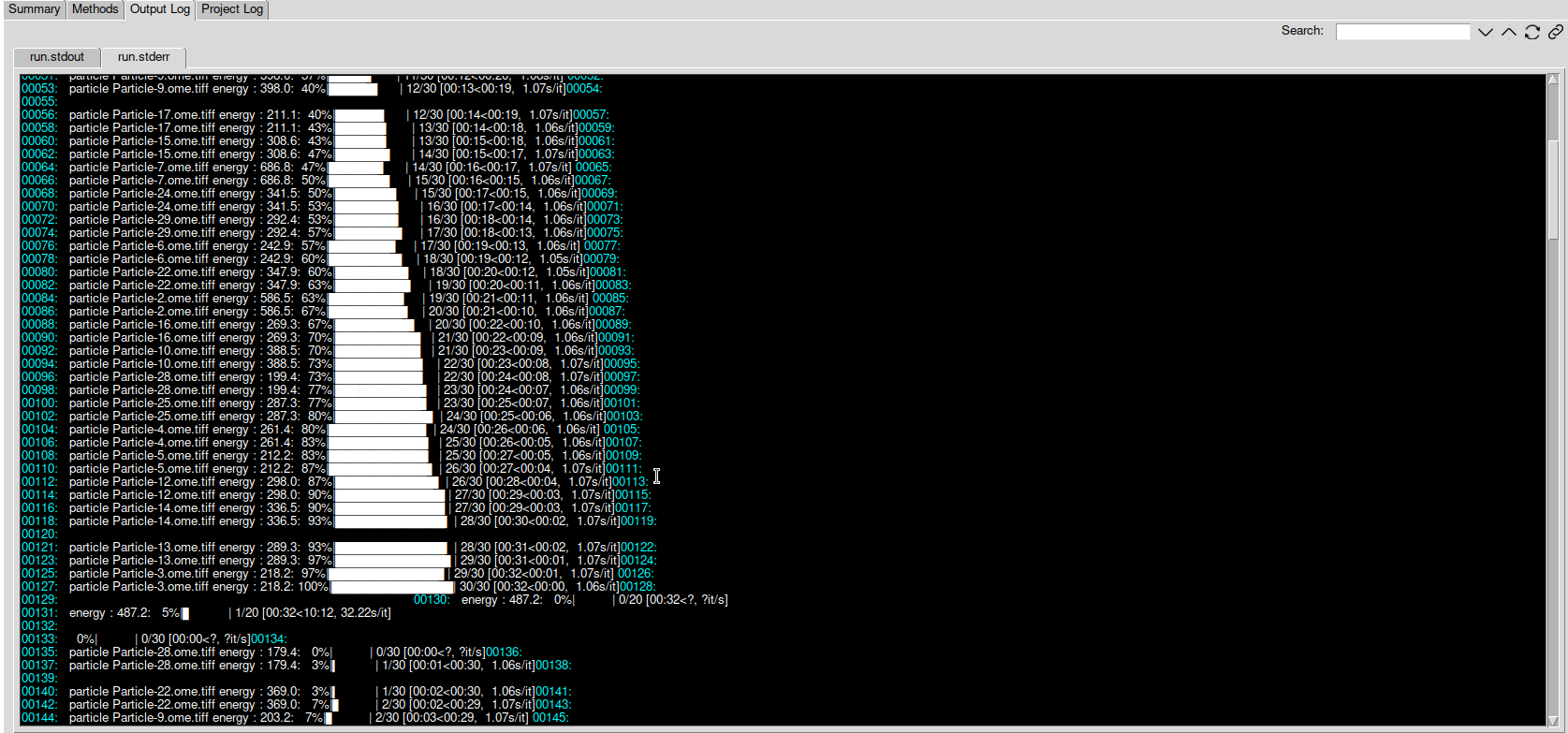
On line 90, you can see:
00090: particle Particle-16.ome.tiff energy : 269.3: 70%|███████ | 21/30 [00:22<00:09, 1.06s/it]
which reads: Particle-16 has an energy of 269.3. It is the 21th particle processed in the epoch, 9 particles are left. The epoch has been running for 22s and will end in approximately 9s.
On line 131, you can see:
00131: energy : 487.2: 5%|▌ | 1/20 [00:32<10:12, 32.22s/it]
which reads: The mean energy for the 1st epoch is 487.2. The epoch was done in 32s. The rest of the epochs will be done in approximately 10m12s.
- When the protocol is finished, you will have 2 outputs:
a reconstructed volume that you can visualise
a set of particles with approximated rotations. Here the visualisation is the same as the raw extracted particles, but internally each particle stores a rotation found during the ab initio reconstruction. They will be used in the
refinementprotocol.
The reconstructed volume might not be aligned with the vertical axis. To impose a cylindrical symmetry constraint during the refinement, we first need to align the volume with the align axis protocol!
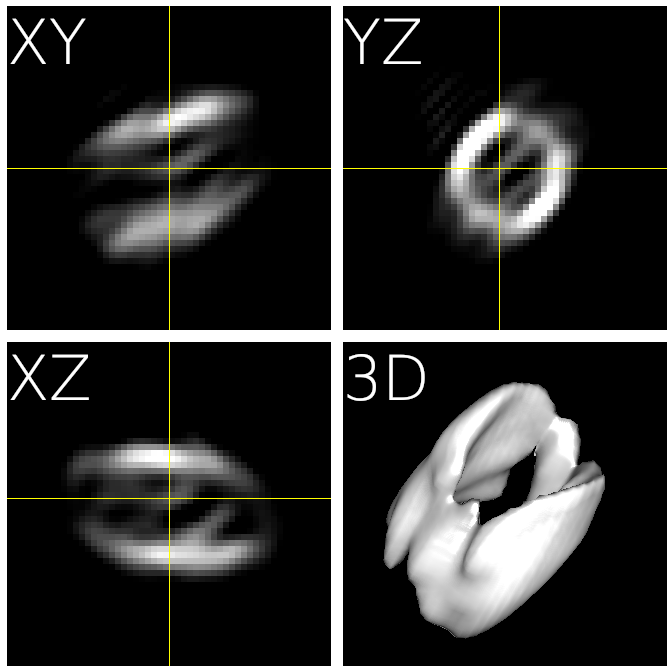
Fig. 2 The ab initio reconstruction you should get¶